How fast is ccs?
Latest version
ccs v6.0 can process 200 GBases HiFi yield in 24 hours for a 25 KBases library on 2x64 cores at 2.4 GHz. To put this into perspective for actual sequencing collections:
| Sample | Insert size | HiFi Yield | Run Time |
|---|---|---|---|
| HG002 | 15 KBases | 41.1 GBases | 4h 49m |
| HG002 | 18 KBases | 26.1 GBases | 2h 53m |
| Redwood | 25 KBases | 32.4 GBases | 3h 17m |
Relative performance v3.0 to v6.0
Current ccs v6 achieves a >150x speed-up for 20 KBases inserts compared to v3.0 from SMRT Link 6.0 release in 2018.
Algorithmic complexity
To understand how this performance gain was possible, an overview how we changed the algorithmic complexity and how ccs scales with insert size and number of passes:
| CCS version | O(insert size) | O(#passes) |
|---|---|---|
| ≤3.0.0 | quadratic | linear |
| 3.4.1 | linear | linear |
| ≥4.0.0 | linear | sublinear |
To visualize this table, we benchmarked runtime using 500 ZMWs per length bin with exactly 7 passes.
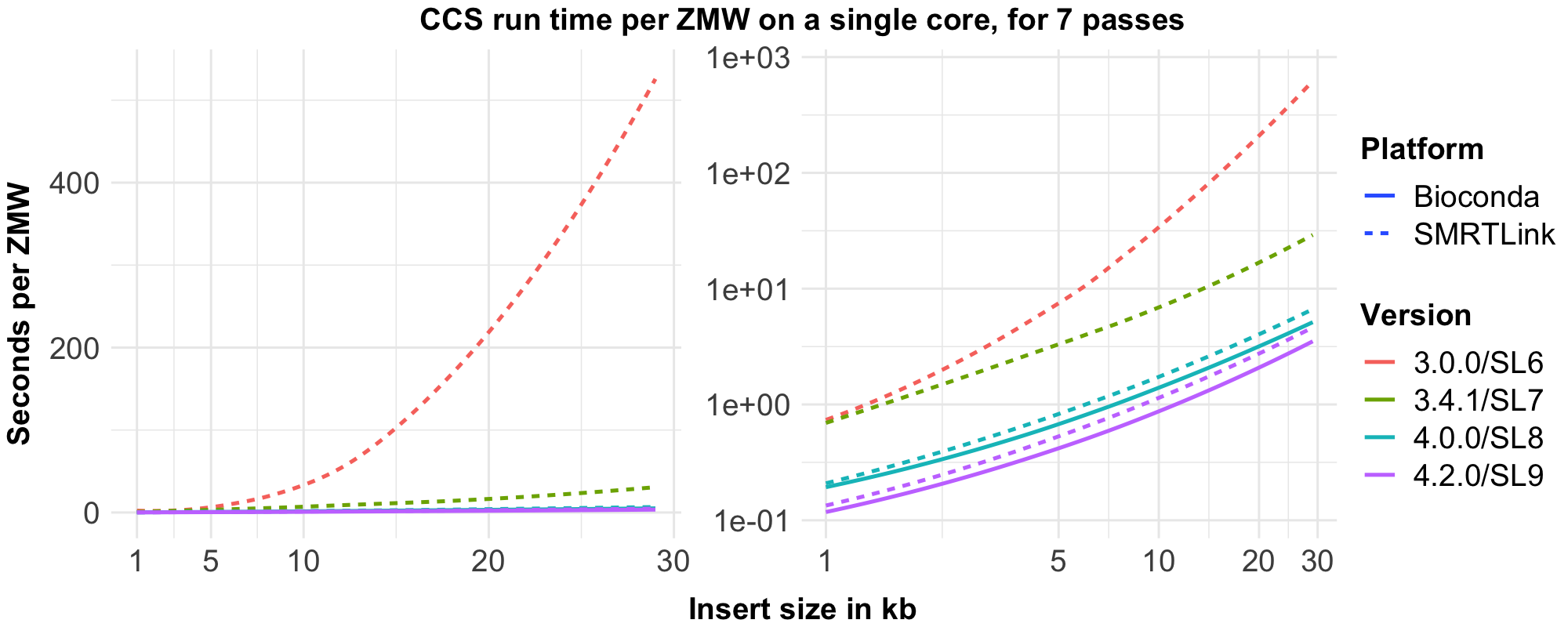
After v4.0.0, the slope of the curve does not change, as the complexity class hasn’t changed; only improvements independent of input type were made.
Performance comparisons
Performance comparisons on different libraries; the faster column is with respect to the run time of the previous version. All runs were performed on the same hardware with 256 threads. A major part of the speed increase in v5.0 is due to toolchain improvements for generating a more optimized binary.
HG002 15kb SQII, 41 GBases HiFi yield
| CCS Version | HiFi Reads | Run Time | CPU Time | Peak RSS | Faster |
|---|---|---|---|---|---|
| 4.0.0 | 2,765,431 | 13h 14m | 89d 13h | 71 GB | |
| 4.2.0 | 2,806,886 | 10h 47m | 61d 9h | 72 GB | 18% |
| 5.0.0 | 2,807,317 | 6h 44m | 62d 22h | 27 GB | 37% |
| 6.0.0 | 2,831,192 | 5h 52m | 44d 17h | 20 GB | 13% |
| 6.2.0 | 2,832,543 | 4h 49m | 50d 19h | 26 GB | 18% |
HG002 18kb SQII, 26 GBases HiFi yield
Omitting v4.0.0, due to lack of chemistry support.
| CCS Version | HiFi Reads | Run Time | CPU Time | Peak RSS | Faster |
|---|---|---|---|---|---|
| 4.2.0 | 1,418,685 | 5h 39m | 54d 14h | 28 GB | |
| 5.0.0 | 1,419,372 | 4h 43m | 42d 18h | 48 GB | 17% |
| 6.0.0 | 1,432,826 | 3h 28m | 28d 17h | 19 GB | 26% |
| 6.2.0 | 1,433,066 | 2h 53m | 30d 9h | 23 GB | 17% |
Redwood 25kb SQII, 32 GBases HiFi yield
| CCS Version | HiFi Reads | Run Time | CPU Time | Peak RSS | Faster |
|---|---|---|---|---|---|
| 4.0.0 | 1,269,680 | 7h 58m | 60d 19h | 72 GB | |
| 4.2.0 | 1,310,775 | 6h 37m | 43d 18h | 74 GB | 17% |
| 5.0.0 | 1,311,693 | 4h 36m | 41d 13h | 41 GB | 30% |
| 6.0.0 | 1,335,888 | 3h 56m | 25d 11h | 17 GB | 14% |
| 6.2.0 | 1,335,674 | 3h 17m | 31d 15h | 22 GB | 18% |
How is CCS speed affected by raw base yield?
Raw base yield is the sum of all polymerase read lengths. A polymerase read consists of all subreads concatenated with SMRTbell adapters in between.
Raw base yield can be increased with 1) higher percentage of single-loaded ZMWs and 2) longer movie times that lead to longer polymerase read lengths.
Since the first version, ccs scales linear in (1) the number of single loaded ZMWs per SMRT Cell. Starting with version 3.3.0 ccs scales linear in (2) the polymerase read length and with version 4.0.0 ccs scales sublinear.
How can version 4.0.0 be sublinear in the number of passes?
With the introduction of improved heuristics, individual draft bases can skip polishing if they are of sufficient quality. The more passes a ZMW has, the fewer bases need additional polishing.
Can I tune ccs to get improved results?
No, we optimized ccs such that there is a good balance between speed and output quality.
Does speed impact quality and yield?
Yes it does. With >150x speed improvements from version 3.0 to 6.0, heuristics and changes in algorithms lead to slightly different yield and accuracy if run head-to-head on the same data set. Internal tests show that ccs 6.0 introduces no regressions in ccs-only Structural Variant calling and has minimal impact on SNV and indel calling in DeepVariant. In contrast, lower DNA quality and sample preparation has a bigger impact on quality and yield.
Can I tune performance without sacrificing output quality?
The bioconda ccs ≥v5.0 binaries statically link mimalloc. Depending on your system, additional performance tuning can be achieved. Internally, we use following mimalloc environment variables to improve ccs performance.
MIMALLOC_PAGE_RESET=0 MIMALLOC_LARGE_OS_PAGES=1 ccs <movie>.subreads.bam ccs.bam --log-level INFO